Tutorial
- Where do I begin?
- Where does the data come from?
- Description of the proteins
- Phosphorylation and kinase information
- How to optimally use advanced search options?
- How to extract subnetworks for proteins of interest and download files to visualize in Cytoscape?
- Images not printing when trying to print. What do I do?
- Still having questions! What do I do?
3. Description of the proteins
Once the protein's name is entered in the search field, a list of proteins matching the search term appears. By clicking on the protein's name in the list, the user can then open the page containing its description. This page includes the information about the protein main name, its multiple identifiers, the study and the fraction where the protein has been found, its phosphorylation sites and alternative names. It also contains additional protein characteristics such as motifs, domains, Gene Ontology (GO) terms and predictions for transmembrane domains along with drugs and human diseases associated with the protein.
In the special case of kinases, it is possible to examine all the phosphorylation targets of the specific kinase and link to the targets' description directly.
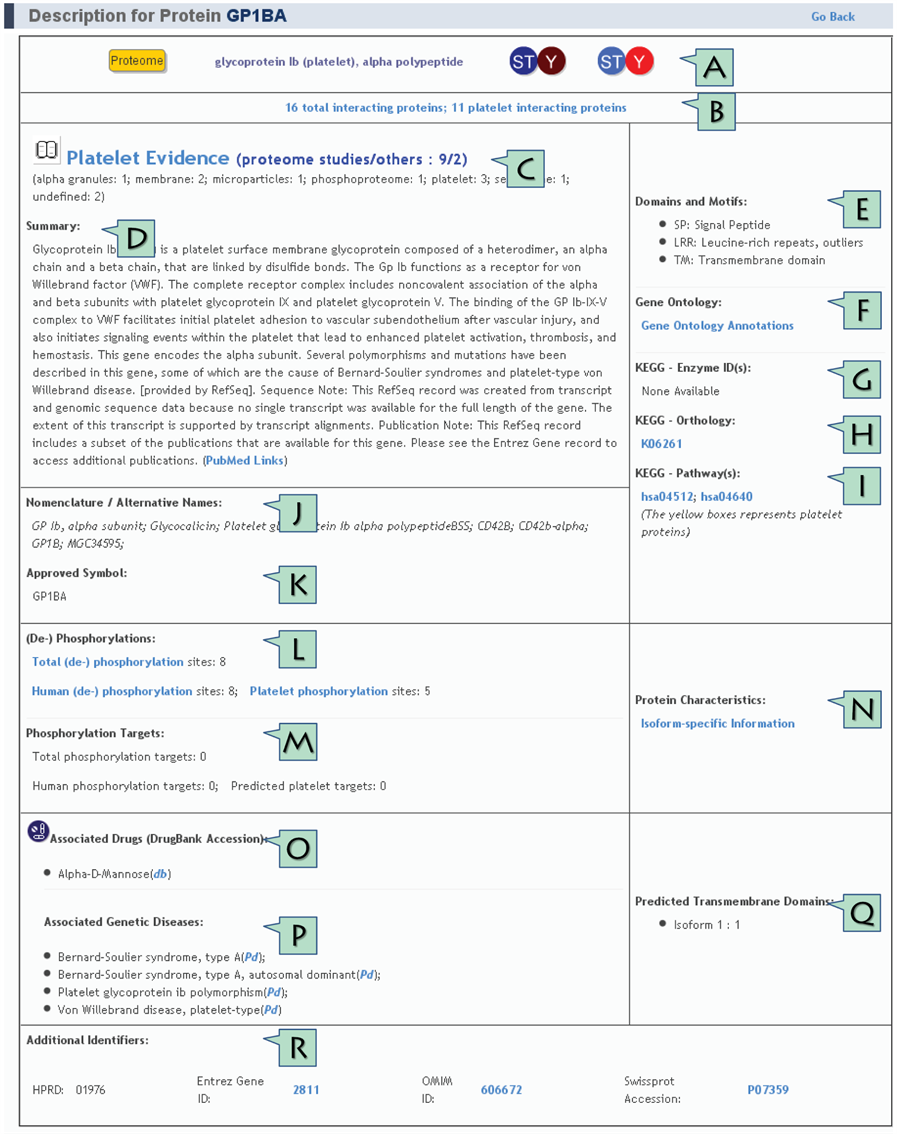 |
A. General information about the protein |
||||
| Refer to the legend for the complete information | ||||
B. Total Interacting Proteins / Platelet Interacting Proteins |
||||
| Exclusive information on the total interacting proteins and the platelet interacting proteins. | ||||
| Total interactions | Clicking on "total interacting proteins" retrieves the list of all protein interactions. | |||
| Platelet interactions | Clicking on "platelet interacting proteins" retrieves the list of interactions with platelet proteins. | |||
| Back to Top | ||||
C. Platelet Evidence |
||||
| Clicking on this link would provide a list of proteome studies, in which the protein was detected.
Furthermore, it contains information about the cellular compartment of the protein along with a link to the study. The studies are divided into 2 groups, proteome data and database/SAGE source information.
Additionally, the studies are categorized according to the cell fraction of the protein and the number of appearances in each fraction is displayed. |
||||
| Back to Top | ||||
D. Summary |
||||
| A short description of the protein and its functions is presented along with the links to PubMed. | ||||
| Back to Top | ||||
E. Domains and Motifs |
||||
| All domains and motifs of the protein are listed with a short abbreviation of the domain name and a complete domain description where available. | ||||
| Back to Top | ||||
F. Gene Ontology |
||||
| Most human proteins have a specific GO-annotation, containing information on the cellular components, biological processes and molecular functions associated with it. Clicking on the link
provides full listing of all the GO annotations of the protein. Further clickable are the GO terms, which contain at least 1 but not more than 150 platelet proteins. A link to all related children terms is displayed next to each of the protein's GO terms. |
||||
| Back to Top | ||||
G. KEGG - Enzyme ID(s) |
||||
| This will provide an external link to the KEGG enzyme information if the protein is a known enzyme. | ||||
| Back to Top | ||||
H. KEGG - Orthology |
||||
| This will provide an external link to the KEGG orthology information of the protein. | ||||
| Back to Top | ||||
I. KEGG Pathways |
||||
| All KEGG pathways, in which the protein is found, are listed and platelet proteins are highlighted in yellow. The visualization of pathways was created using the software Advanced Pathway Painter. Displayed protein names are not necessarily the HUGO approved symbols, as they are mapped automatically to KEGG pathways' nomenclature in this software. | ||||
| Back to Top | ||||
J. Nomenclature / Alternative Names |
||||
| Other names assigned to the protein. | ||||
| Back to Top | ||||
K. Approved Symbol |
||||
| This is the HUGO approved Symbol of the protein. | ||||
| Back to Top | ||||
L. (De-) Phosphorylations |
||||
| Total phosphorylations, divided into human phosphorylation sites and platelet phosphorylation sites. Clicking on the "(De-) Phosphorylations" link retrieves a list of all phosphorylated and de-phosphorylated residues and their position in the protein's sequence. It is also possible to view the human (de-) phosphorylations and platelet phosphorylations individually. Clicking on any of these links will navigate to the next page where each phosphosite is displayed with its residue and site. Underneath this information, you can find the isoforms in which the site is identified. Further down within the same phosphosite, one can find the phosphorylated motif sequence with the site highlighted in the middle. Additionally, the phosphorylating kinase (if available) is shown next to it. The source of phosphorylation information is presented under Reference. For kinase predictions on platelet phosphorylation sites the reference is given as NetworKIN (prediction algorithm). For all data from literature, the reference is either PhosphoSite or HPRD (Source: PubMed). Finally, each phosphorylation is presented with the type of experiment used for its detection. |
||||
| Back to Top | ||||
M. Phosphorylation Targets |
||||
| This is only valid if the protein is a kinase. Clicking on the "Phosphorylation Targets" link retrieves a list of all proteins phosphorylated by the kinase. It is also possible to view the "human phosphorylation targets" and "predicted phosphorylation targets" individually. |
||||
| Back to Top | ||||
N. Protein Characteristics |
||||
| It gives isoform-specific information about the physical properties of the protein. When clicked, details on the protein's molecular weight, isoform accession, its length, isoelectric point and the protein sequence in a downloadable FASTA format are presented. | ||||
| Back to Top | ||||
O. Associated Drugs (DrugBank Accession) |
||||
| All drugs are listed with their corresponding accession number for DrugBank. Clicking on the link "(db)" retrieves the DrugBank webpage with detailed information about the drug. | ||||
| Back to Top | ||||
P. Associated Genetic Diseases |
||||
| Clicking on the "(Pd)" link next to each genetic disease term navigates to the pubmed information source. | ||||
| Back to Top | ||||
Q. Predicted Transmembrane Domains |
||||
| Information on the number of predicted transmembrane domains for each isoform of the protein is shown here. | ||||
| Back to Top | ||||
R. Additional Identifiers |
||||
| External links associated to the protein - identifiers from HPRD, NCBI gene id, OMIM id and the Swissprot accession number are provided here. | ||||
| Back to Top | ||||